
RadBench Dataset#
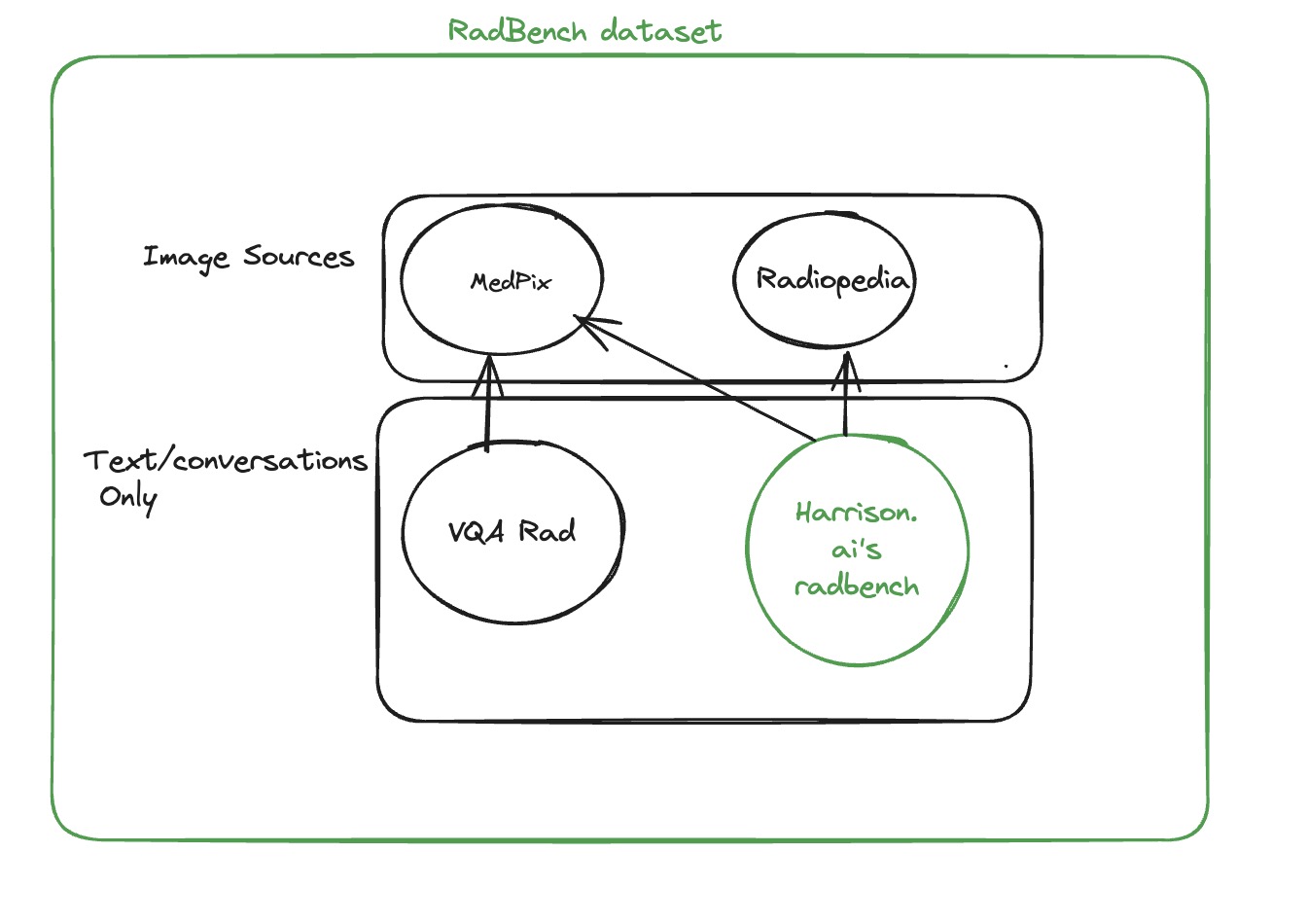
RadBench dataset is collation of clinically relevant Radiology specific visual questions and answers (VQA) based on plain film X-ray. This VQA dataset is clinically comprehensive, covering 3 or more questions per medical imaging. The radiology images for this set are sourced from Medpix and Radiopaedia. RadBench is curated by medical doctors with expertise in relevant fields who interpret these images as part of their clinical duties.
Overview#
The RadBench dataset is formatted similarly to VQA-Rad1 to ensure ease of use by Medical/Radiology communities. Some key differences are:
- Rich set of possible answers: The closed questions of the RadBench dataset have a set of possible answers explicitly defined.
- Level of correctness: The set of possible answers for given question is also ordered in terms of relative correctness. This is done to account for the fact that some options can be more incorrect than others. This ordering also helps with differential diagnosis.
- Multi-turn Questionnaire: Questions are ordered per case by specificity - meaning that if evaluated in the same context, they should be asked in that order. For example, "Is there a fracture in the study?" should be asked prior to "Which side is the fracture on?" as the second question implies the answer to the first.
Why RadBench?#
There has been a growing concern within computer vision and deep learning (CV & DL) communities that we have started to overfit popular existing benchmarks, such as ImageNet 2. We share this concern and worry that Radiology foundation models perhaps are also starting to overfit on VQA-Rad 1. Besides, existing Radiology VQA datasets have several shortcomings:
- Some datasets have automatically generated questions and answers from existing noisy labels extracted from radiology reports. This leads to unnatural and ambiguous questions which cannot be adequately answered given the image. For instance:
- This question
In the given Chest X-Ray, is cardiomegaly present in the upper? (please answer yes/no)(dataset source: ProbMed)3 is anatomically impossible to answer as cardiomegaly is not divided intoupperandlower. - Likewise, in SLAKE 4 dataset, given the image
xmlab470/source.jpg, questionWhere is the brain non-enhancing tumor?is asked. However the image is an axial non-contrast T2 MRI of the brain whereby answering the question of 'non-enhancing tumor' is not possible. The answer for this question is also given asUpper Left Lobewhich is not a valid anatomical region in the brain. This should be answered asanterior left frontal lobe.
- This question
- Some existing datasets have been curated by non-medical specialists, leading to questions which may be less relevant to everyday clinical work and pathology.
- Existing datasets do not include more than one image per question, whereas in radiology many studies do include more than one view. Having only one image does not allow us to evaluate the model for its ability of comparing multiple images at once, which is a very clinically relevant task.
- Existing datasets do not specify the context in which the images should be used. This is relevant to RadBench as more than one image can be used in a single question. In RadBench, the
<i>token is used to denote the location of an image in relation to the surrounding words (more specifically tokens). This allows specific references to the images in the question e.g. "the first study" or "the second study". As a result multi-turn comparison questions can now be asked. - Existing datasets are not selected for clinically challenging cases where the pathology is visually subtle or rare. RadBench specifically selects a wide range of pathology in different anatomical parts with the intention of including challenging cases.
Working with RadBench Dataset#
The RadBench dataset is a collection of 89 unique cases, consisting of 40 cases sourced from MedPix and 49 cases sourced from Radiopaedia. A total of 497 questions were asked for these cases, with 377 questions being closed-ended and 120 questions being open-ended.
Here is a breakdown of the dataset's structure:
| Header | Description |
|---|---|
| imageSource | Describes the source of the images, indicating whether they are from MedPix or Radiopaedia. |
| CASE_ID | For MedPix cases, this field is left blank. For Radiopaedia cases, it contains a numerical value that can be used to search for the case image at Radiopaedia.org using the provided case ID. |
| imageIDs | A list of unique IDs for each image, separated by commas. These IDs can be either MedPix image IDs or Radiopaedia image URLs. To obtain URLs for MedPix images, refer to the following section on loading RadBench images. |
| modality | Currently, the dataset only includes "XR - Plain Film" modality. |
| IMAGE_ORGAN | Specifies the organ present in the image, such as ABDOMEN, BRAIN, or PELVIS. |
| PRIMARY_DX | Indicates the primary diagnosis for the given case. |
| QUESTION | Represents the question to be provided to the AI model. |
| Q_TYPE | Specifies the type of question asked, such as Pathology, Radiological View, Body Part, Clinical, Demographic, or Comparison. |
| ANSWER | Denotes the expected answer from the model. |
| A_TYPE | Indicates whether the question is open-ended or closed-ended. |
| OPTIONS | For closed-ended questions, this field contains a list of answer options, such as "left" and "right". For open-ended questions, this field is left blank. |
Loading RadBench images#
To load the images for the RadBench dataset, you will need to identify the source of the images using the imageSource column provided in the RadBench dataset.
If the images are sourced from Radiopaedia, the image URLs can be found in the imageIDs column and can be directly used to fetch the images.
For MedPix images, you will need to query the MedPix image metadata first and obtain the images. To query the metadata, you can use the following URL structure: https://medpix.nlm.nih.gov/rest/image.json?imageID=. Simply append the image ID to the end of the URL. The response will be in JSON format, with the key imageURL containing the URL to the actual image.
Acknowledgements#
We thank Medpix and Radiopaedia and their respective editorial teams and contributors specially NIH, Frank Gaillard, Andrew Dixon, and other Radiopaedia.org contributors for creating such a rich library of cases to test radiology expertise.
-
JJ Lau, S Gayen, Abacha A Ben, and D. Demner-Fushman. A dataset of clinically generated visual questions and answers about radiology images. Scientific Data, 5(1):2052–4463, 2018. URL: https://www.nature.com/articles/sdata2018251. ↩↩
-
Lucas Beyer, Olivier J. Hénaff, Alexander Kolesnikov, Xiaohua Zhai, and Aäron van den Oord. Are we done with imagenet? CoRR, 2020. URL: https://arxiv.org/abs/2006.07159. ↩
-
Q. Yan, X. He, X. Yue, and X. E. Wang. Worse than random? an embarrassingly simple probing evaluation of large multimodal models in medical vqa. ArXiv, 2024. URL: https://arxiv.org/abs/2405.20421. ↩
-
B. Liu, L. Zhan, L. Xu, L. Ma, Y. Yang, and X. Wu. Slake: a semantically-labeled knowledge-enhanced dataset for medical visual question answering. ArXiv, 2021. URL: https://arxiv.org/abs/2102.09542. ↩